Amplification of the barcode sequencing
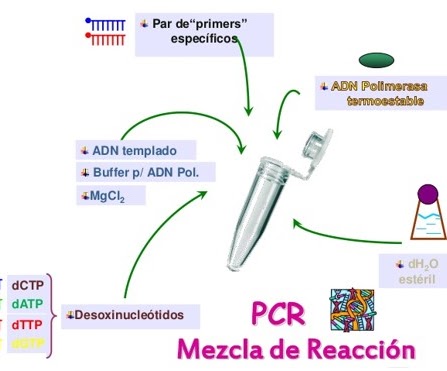
Prepare a mixture of reagents to detect and copy the selected sequence as a target in the DNA of the microorganisms:
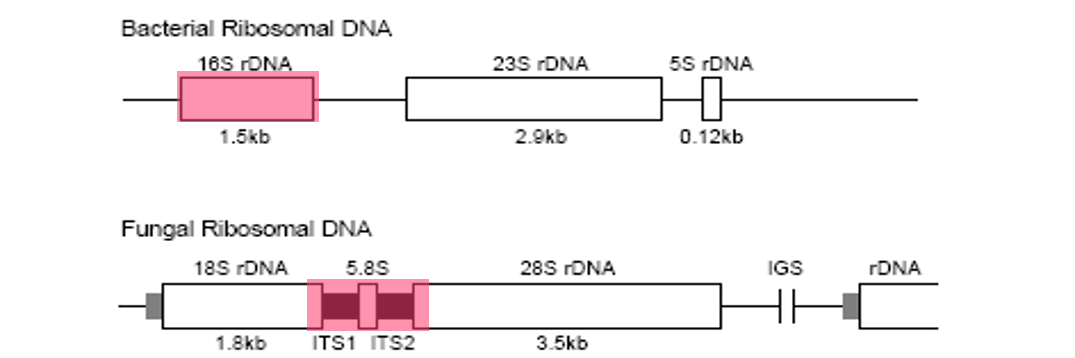
– with regard to the bacteria, the target will be a fragment of DNA that is encoded for the small ribosomal subunit (16S).
– as for the yeast, a fragment that includes the internal transcribed spacer (ITS) regions that exist between the gene that is encoded for the small subunit 18S and the large subunit 28S, which in turn includes the small subunit 5.8S.
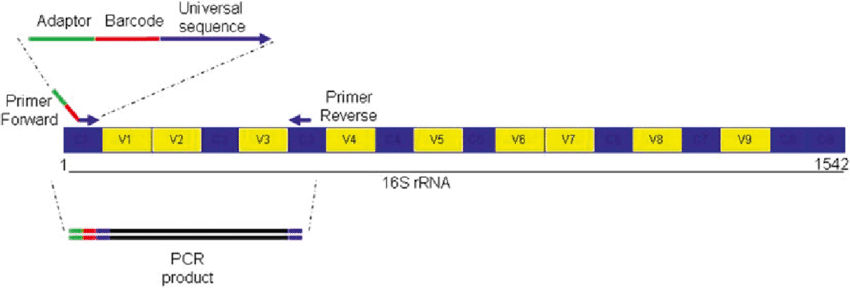
In this mixture it will only be different in the primers, which are those that must be specifically joined to the target regions. The other elements are the same. To make the mixture the calculation is based on a reagent volume of 25 μL for each sample, which means that for N samples the composition will be the one that is shown in the figure.
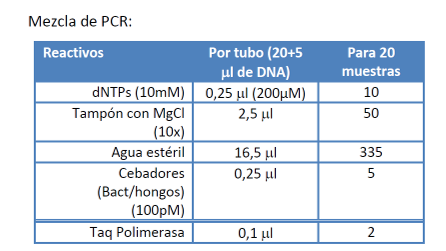
Gently mix the reaction components and divide 20 microlitres into PCR tubes.
Add 5 microlitres of extracted DNA to each sample, taking into account whether it is bacterial or fungal DNA, for amplification with the corresponding primers.
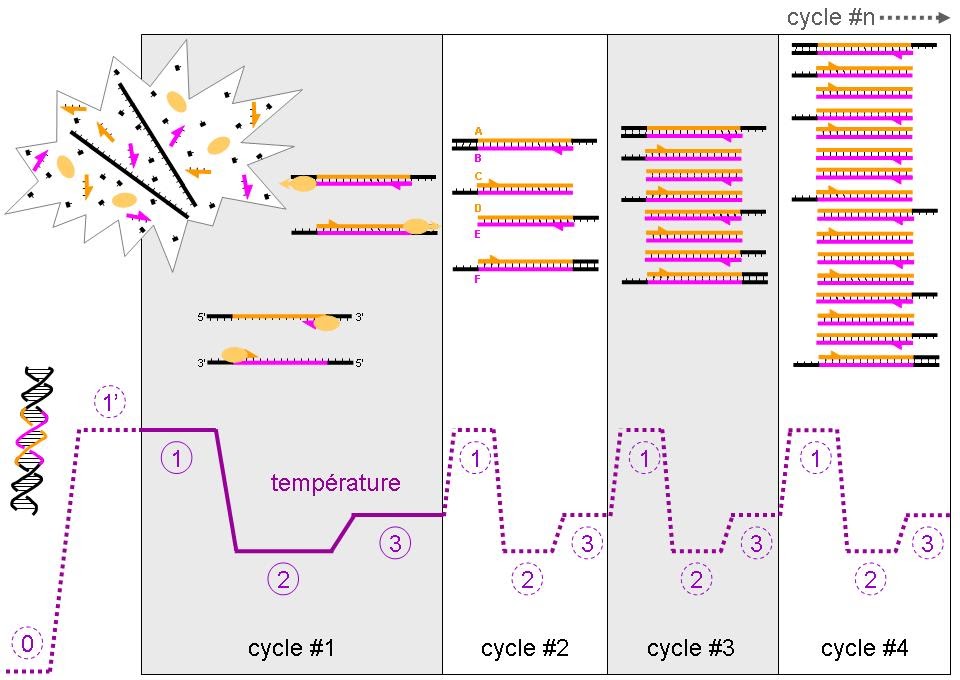
Transfer to the thermal cycler with the following programme:
1 cycle: 95ºC ………….5 min
35 cycle: 95ºC………30 sg
55ºC……30 sg
72ºC ….. 1 min
1 cycle: 72ºC ,,,,,,,6 min
Apoyo a acciones de Innovación Docente – Vicerrectorado de Estudios / Consultas e incidencias técnicas- Tlf: 96 522 2059 –